New paper in PNAS
Holographic virtual staining of individual biological cells
via Deep Learning
Y. N. Nygate, M. Levi, S. Mirsky, N. A. Turko, M. Rubin, I. Barnea, M. Haifler, A. Shalev,
and N. T. Shaked
Proceedings of the National Academy of Sciences of the U.S.A. (PNAS),
Vol. 117, No. 17, pp. 9223-9231, 2020
Abstract: Many medical and biological protocols for analyzing individual biological cells involve morphological evaluation based on cell staining, designed to enhance imaging contrast and enable clinicians and biologists to differentiate between various cell organelles. However, cell staining is not always allowed in certain medical procedures. In other cases, staining may be time consuming or expensive to implement. Here, we present a new deep-learning approach, called HoloStain, which converts images of isolated biological cells acquired without staining by holographic microscopy to their virtually stained images. We demonstrate this approach for human sperm cells, as there is a well-established protocol and global standardization for characterizing the morphology of stained human sperm cells for fertility evaluation, but, on the other hand, staining might be cytotoxic and thus is not allowed during human in vitro fertilization (IVF). We use deep convolutional Generative Adversarial Networks (DCGANs) with training that is based on both the quantitative phase images and two gradient phase images, all extracted from the digital holograms of the stain-free cells, with the ground truth of bright-field images of the same cells that subsequently underwent chemical staining. To validate the quality of our virtual staining approach, an experienced embryologist analyzed the unstained cells, the virtually stained cells, and the chemically stained sperm cells several times in a blinded and randomized manner. We obtained a 5-fold recall (sensitivity) improvement in the analysis results. With the introduction of simple holographic imaging methods in clinical settings, the proposed method has a great potential to become a common practice in human IVF procedures, as well as to significantly simplify and facilitate other cell analyses and techniques such as imaging flow cytometry.
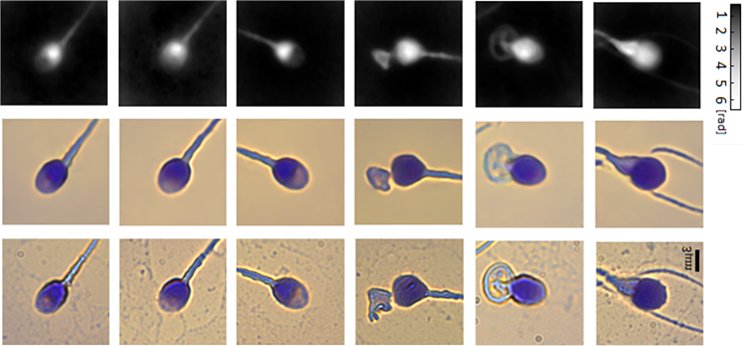
Virtual staining of individual sperm cells using single-projection interferometry. We used the label-free free quantitative phase profiles of the cells as inputs to a neural network that performed the mapping, after being trained with the coinciding chemical staining images. The first row shows the quantitative phase images extracted from the single holograms. The second row shows the coinciding virtual stained images, generated by the network. The third row shows the coinciding bright-field chemically stained images of the same sperm cells that the network did not see; yet it can present the cells as if they were stained, as shown in the second row, based only on the label-free quantitative phase images shown in the first row. The first three columns show normal morphology cells, and the last three columns show pathological cells.